Say you started working on this data set and you are planning to build a classification model(for the sake of discussion). You did data munging (cleaning, feature engineering, etc.) and you are now ready to split the data for training and testing. For the sake of simplicity, let us assume you have two classes in your data, A and B, with 70 of total data belonging to class A and rest to B. When you divide the data into ratio of train and test data, random number of samples of class A and B will flow to train and test data(while maintaining the size ratio of train and test as 4:1). What if 65 of data belonging to class A and 5 of class B goes to training set. Training set will be dominated with samples from class A (65) while test set will only have (5). Since our training set has majority of data labeled as class ‘A’, 93% to be precise, model tries to overfit resulting in high variance. When the same model is applied to testing data, which has 25 records labeled as ‘B’, they will be classified as ‘A’ resulting in horrible test score. This is because that training and testing data are not homogeneous.
TL;DR, you split the data wrong and now your model is performing poorly because of high variance. Root cause of this is that we did not take percentages of labels into consideration during the split. Ideally we should same distribution of labels in test/validation set as the distribution in training set.
Importance of Validation set
The above mentioned scenario is just one such case where model performs well on training set and does badly on validation/testing set. Overfit or underfit can also occur when we chose an imperfect model to fit data. An imperfect model is something that is designed to work well on the kind of data (like decision trees on categorial features) we have, although it can perform well on other data sets. This happens because algorithms are built based on few assumptions on data formats and are not supposed to fit other kinds of data equally well.
We need to have some early indicator informing if the model is overfitting. This is where validation accuracy comes into picture and help us identify high variance before its too late. Low accuracy on validation set upon fitting the model on it is an indicator of high variance in the model and it’s time we either go for a different mode or re-engineer the data. We need a model that’s good, not the one that looks good.
Single vs Multiple validation sets
It has been shown by several researchers that validation using single test can be a poor choice. Resampling methods, such as cross-validation, can be used to produce appropriate estimates of model performance using the training set. Resampling methods often produce performance estimates superior to a single test set because they evaluate many alternate versions of data.
Different ways to get CV data sets
sklearn’s modelselection class has various subclasses which can do the data partitions very effectively. In case of supervised learning where the test
data is not provided, the data should be split into three sets for training, validation and testing purpose. If the test set already exists (provided), we only
need to create training and validation sets.
In this article, I shall briefly discuss about some the commonly used resampling techniques offered by sklearn and will explain their pros and cons. There are three major kinds of splits, K-Fold, stratified and shuffle split.
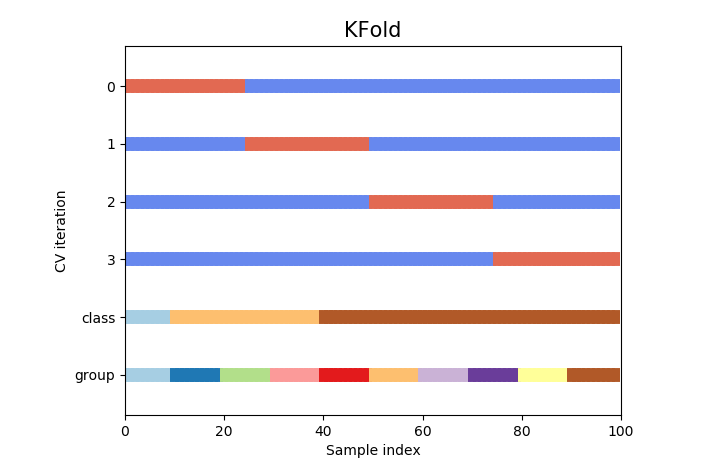
K-fold Cross-Validation
- The samples are randomly partitioned into k sets of roughly equal size
- Indexes of data points in subsamples follow same order as that in original data
- The k resampled estimates of performance are summarized (usually with the mean and standard error) and used to understand the relationship between the tuning parameter(s) and model utility
- The choice of k is usually 5 or 10, but there is no formal rule
- Does not take percentage of data in the classes into consideration during split
from sklearn.model_selection import KFold
n_points = 100
n_splits = 4
X = np.random.randn(100, 10)
"""
Create sample data with 3 classes and 10 groups (Each sized 10)
"""
percentiles_classes = [.1, .3, .6]
y = np.hstack([[ii] * int(100 * perc)
for ii, perc in enumerate(percentiles_classes)])
# Evenly spaced groups repeated once
groups = np.hstack([[ii] * 10 for ii in range(10)])
#create CV object and split the data
cv = KFold(n_splits)
cv.split(X=X,y=y, groups=groups)
The obvious pitfall of this way of splitting is that it does not take into consideration the population of various target classes in the data. This could lead to one or more classes not appearing in training set thus enabling model to overfit on labels appearing in training set.
To overcome this we need have a resampling technique that helps us to split the data according to the percentage of classes so that we have it evenly distributed in training and validation data. Stratified split helps us to achieve that.
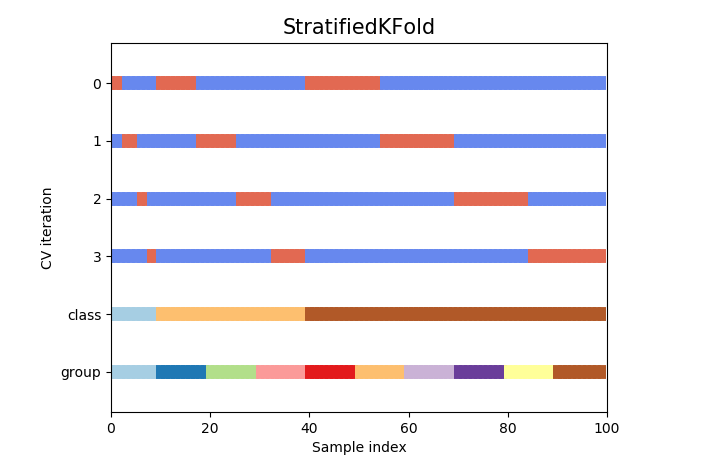
Stratified split
As mentioned above, stratified split(stratifiedKFold split to be precise) splits the data by keeping distribution of target variable into consideration.
- Enhanced K-fold split where split is done while preserving the percentage of samples for each class
# Create cross-validation object
cv_strat_split = StratifiedKFold(n_splits)
cv_strat_split.split(X=X,y=y, groups=groups)
There is still the issue of splitting data linearly. It would be better if we can somehow split the data while preserving the population of classes in training and validation sets.
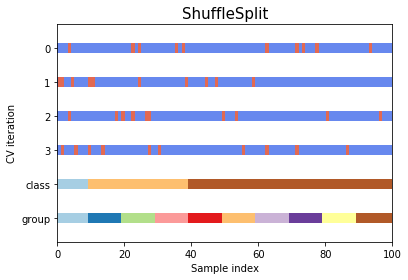
Shuffle split
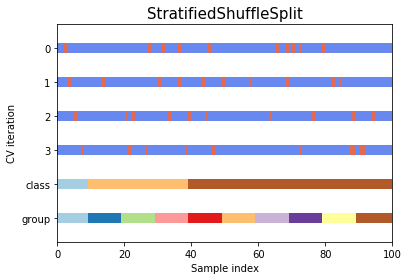
Shuffle splitsplits the by randomly picking indexes from pool of available data (not in order)Stratified shuffle splitshuffles the data like above while preserving the percentage of classes in the distributed data- Stratified shuffle split is particularly helpful if there is large disparity between the population of classes in the dataset
#Shuffle split
cv_shuff_split = ShuffleSplit(n_splits, test_size=.25, random_state=0)
cv_shuff_split.split(X=x)
#Stratified shuffle split
cv_start_shuff = StratifiedShuffleSplit(n_splits, test_size=0.25, random_state=0)
cv_start_shuff.split(X=x)

Leave a Comment